After I wrote about Extracting RDF data models from Wikidata in my blog last month, Ettore Rizza suggested that I check out wdtaxonomy, which extracts taxonomies from Wikidata by retrieving the kinds of data that my blog entry's sample queries retrieved, and it then displays the results as a tree. After playing with it, I'm tempted to tell everyone who read that blog entry to ignore the example queries I included, because you can learn a lot more from wdtaxonomy.
The queries in that blog entry might still give you some useful perspective on how SPARQL can retrieve triples from Wikidata that express tree-ish relationships between the concepts of a given domain that have Wikipedia pages--whether you want to call that a taxonomy or an ontology--but I was just dabbling, while wdtaxonomy is a full-featured serious application for this.
Jakob Voss designed wdtaxonomy as both a command line utility and as an NPM module that you can reference from applications. I tried the command line version and had a lot of fun. To try it with my periodic table element example that I wrote about last month, I started by entering "wdtaxonomy Q11344" (using the same local name for the Wikidata identifier that I used before) and the results were impressive.
wdtaxonomy typically outputs a text-based tree with various information about the nodes of the tree. Instead of pasting a sample here, I'm showing a screen shot of the beginning of the output so that you can see the nice color coding:
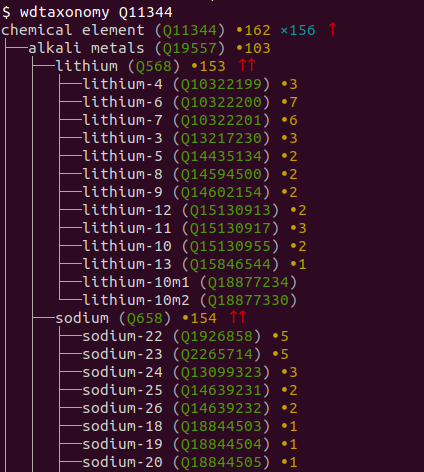
The wdtaxonomy readthedocs.io documentation lists over two dozen command line options that you can use to customize the output. (Entering "wdtaxonomy" alone at the command line gives a good summary.) My favorite is -s, which tells you you the SPARQL query that wdtaxonomy would use to retrieve the requested information from wikidata. Here is what that gives you when added it to the Q11344 command line I entered above:
$ wdtaxonomy -s Q11344
SELECT ?item ?broader ?itemLabel ?instances ?sites WITH {
SELECT DISTINCT ?item { ?item wdt:P279* wd:Q11344 }
} AS %items WHERE {
INCLUDE %items .
OPTIONAL { ?item wdt:P279 ?broader } .
{
SELECT ?item (count(distinct ?element) as ?instances) {
INCLUDE %items.
OPTIONAL { ?element wdt:P31 ?item }
} GROUP BY ?item
}
{
SELECT ?item (count(distinct ?site) as ?sites) {
INCLUDE %items.
OPTIONAL { ?site schema:about ?item }
} GROUP BY ?item
}
SERVICE wikibase:label {
bd:serviceParam wikibase:language "en"
}
}
(The INCLUDE keyword used in this query is a Blazegraph and Anzo extension to the SPARQL standard.) Combining this -s option with other options, such as -i to include instances or -d to include item descriptions, shows what SPARQL query the tool would generate to retrieve this additional information. It's a great opportunity to learn more about SPARQL, about the Wikidata data model, and about their relationship. (I have worried that this data model would scare off people who are new to SPARQL--that if their first data set to query was Wikidata, they migh think that the complexity of the necessary queries was because of SPARQL and not because of Wikidata--but when I see all the great activity on Twitter around the use of SPARQL with Wikidata lately, I don't worry so much anymore.)
The ability to get at the generated SPARQL queries is also a huge help to my original goal of retrieving triples that let me store an RDFS/OWL ontology or a SKOS taxonomy about Wikipedia entities. I can change the SELECT part to a CONSTRUCT clause to create triples that use the variables bound in wdtaxonomy's WHERE clauses. wdtaxonomy (or rather, Jakob) has done the difficult work of assembling the necessary query logic and we can just take it and use it.
Some of the other command line options I liked include -U to get full URIs and -r to get superclasses of the named entity instead of its subclasses. I encourage everyone interested in SPARQL and Wikidata to install wdtaxonomy and start playing with it. Especially with that -s option!
Please add any comments to this Google+ post.

